
PHERAstar FSX
Powerful and most sensitive HTS plate reader
PROTACs are small, readily designed molecules that target unwanted proteins to the cell’s ubiquitin-proteasome system for degradation. Find out how microplate readers can advance PROTAC research.
 Dr Barry Whyte
Dr Barry Whyte
PROteolysis TArgeting Chimeras (PROTACs) are small molecules that selectively target unwanted proteins to the cell’s protein disposal system (ubiquitin-proteasome system). They are designed to selectively degrade disease-causing proteins and present new opportunities for therapeutic intervention. In this blog, we look at the rapidly emerging world of PROTACs: what they are and what they can do. We also discuss how microplate readers can be used to accelerate PROTAC research.
PROteolysis TArgeting Chimeras or PROTACs are an emerging class of molecules that offer new ways to attack disease-causing proteins. They offer the distinct advantage of being able to tackle many previously undruggable targets.1 A PROTAC consists of two separate molecules bound together to form a two-pronged molecular entity (Fig. 1). These two-headed molecules are constructed to selectively degrade disease-causing proteins using the cell’s very own protein disposal machinery, the ubiquitin-proteasome system.
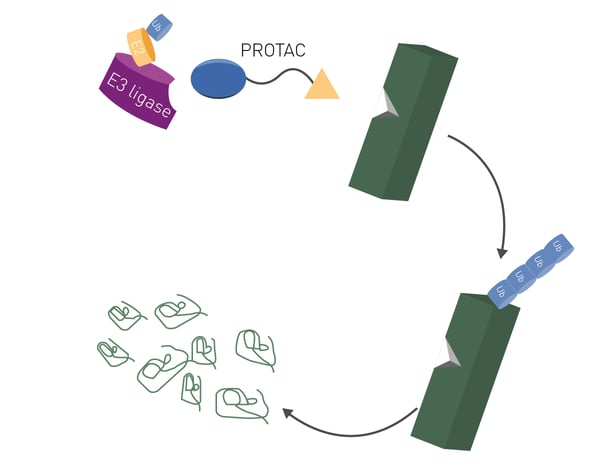 The concept underpinning PROTACs involves a bifunctional molecule that consists of two essential components: a ligand for the target protein and another ligand for an E3 ubiquitin ligase enzyme. The target ligand binds the protein of interest while the E3 ubiquitin ligase ligand recruits an E3 ligase enzyme (Fig. 1). The recruited ligase brings the target protein into proximity to the ubiquitin-proteasome system, the cell’s very own disposal system responsible for protein degradation.
The concept underpinning PROTACs involves a bifunctional molecule that consists of two essential components: a ligand for the target protein and another ligand for an E3 ubiquitin ligase enzyme. The target ligand binds the protein of interest while the E3 ubiquitin ligase ligand recruits an E3 ligase enzyme (Fig. 1). The recruited ligase brings the target protein into proximity to the ubiquitin-proteasome system, the cell’s very own disposal system responsible for protein degradation.
Once the target protein and E3 ligase are brought together, the ubiquitin ligase adds ubiquitin molecules to the target protein marking it for degradation. Once marked, the tagged protein is recognized by the proteasome and degraded into smaller peptides and amino acids. This degradation process takes the disease-causing protein out of circulation (Fig. 2). 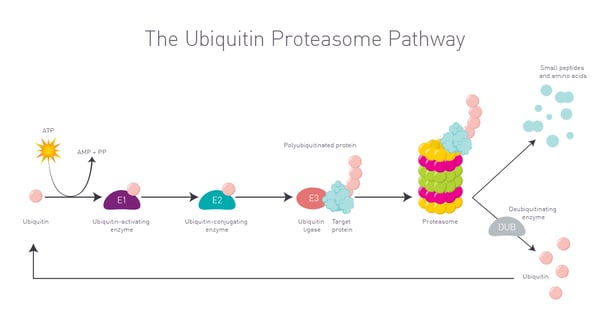 One of the big advantages of PROTACs is their ability to target proteins that were previously classed as undruggable using traditional small molecule inhibitors. Some disease-causing proteins have inaccessible or atypical active sites. In some cases, disease-causing proteins completely lack any enzymatic activity. This limits the design and use of small molecules as drugs. By focusing on protein degradation instead of inhibition, PROTACs offer a new approach to controlling protein levels in the cell. This has the potential to unlock new therapeutic opportunities for a wide range of diseases including cancer, neurodegenerative disorders and autoimmune conditions. The potential for PROTACs as therapeutics is immediately relevant to disease-related non-enzymatic proteins including transcription factors for which small molecule inhibitors have proven to be ineffective.
One of the big advantages of PROTACs is their ability to target proteins that were previously classed as undruggable using traditional small molecule inhibitors. Some disease-causing proteins have inaccessible or atypical active sites. In some cases, disease-causing proteins completely lack any enzymatic activity. This limits the design and use of small molecules as drugs. By focusing on protein degradation instead of inhibition, PROTACs offer a new approach to controlling protein levels in the cell. This has the potential to unlock new therapeutic opportunities for a wide range of diseases including cancer, neurodegenerative disorders and autoimmune conditions. The potential for PROTACs as therapeutics is immediately relevant to disease-related non-enzymatic proteins including transcription factors for which small molecule inhibitors have proven to be ineffective.
The design of proteolysis targeting chimeras involves the careful selection of ligands for the target molecule and the E3 ligase as well as decision making about the linker molecule connecting both functional ligands. Once a target protein is identified, a ligand is chosen that binds to the target protein. The ligand can be designed to target a particular binding pocket or other site on the protein. An E3 ligase enzyme is identified that will transfer ubiquitin molecules to the target protein (ubiquitin ligase). In addition, a ligand is chosen that specifically binds to the E3 ligase. This ligand acts as a recruiter bringing the E3 ligase and the target together.
The ligands for the E3 ligase and the target protein are connected by a linker molecule. The choice of linker depends on the specific target protein and the desired degradation rate.
Examples of linkers include peptides, small molecules (organic compounds), hydrazones and disulfides. Peptide linkers include Glu-Glu, Lys-Lys and Arg-Arg. Alkyl chains, alkynes and azides are often used as small molecules. Hydrazone linkers are commonly used in PROTACs because of their stability and rapid degradation in cells. Disulfide linkers are stable under physiological conditions but offer the advantage of being able to be cleaved in the presence of reducing agents.
With all the parts selected, the designed PROTAC molecule is then synthesized and tested in cellular and animal model systems to evaluate efficacy and selectivity. Iterative optimization may be performed to improve different properties, including affinity for the target protein and ligase, cell permeability, stability and selectivity. Computational modeling, structure-based drug design and high-throughput screening using microplate readers are often used to aid in the rational design of PROTAC molecules. Furthermore, the PROTAC scaffold is also used for the validation of potential target proteins involved in disease. The so called dTAGs are used to induce degradation of target proteins via a gene edited binding site previously introduced into a protein of interest as highlighted in the application note: dTAG protein degradation assay for the targeted degradation of proteins of interest.
The first proof of concept for PROTACs was reported in a landmark study published in 2001 by Craig Crews and colleagues. The study involved demonstration of the efficacy of PROTAC-1 for the targeted degradation of methionine aminopeptidase-2 (MetAP-2). This groundbreaking study provided the first experimental evidence that small molecule-induced protein degradation using PROTACs was feasible and effective.2 The success of PROTAC-1 has spurred further advancements and optimization of PROTAC design, leading to the development of diverse PROTAC molecules targeting different disease-causing proteins.
Many PROTACs have been developed over the past years. ARV-110 is a PROTAC designed to degrade the androgen receptor which is implicated in prostate cancer. It consists of an androgen receptor ligand known as bicalutamide linked to cereblon (CRBN) as the E3 ligase (Fig. 3). 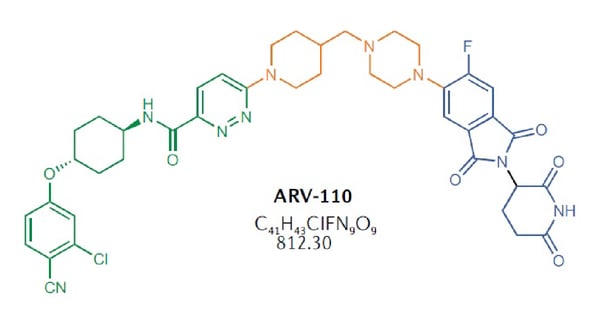 ARV-471 is a PROTAC designed to degrade the estrogen receptor which is involved in certain types of breast cancer. It consists of the receptor ligand fulvestrant linked to cereblon as the E3 ligase. Effective degradation of the estrogen receptor has been demonstrated along with antitumor activity in breast cancer model systems.
ARV-471 is a PROTAC designed to degrade the estrogen receptor which is involved in certain types of breast cancer. It consists of the receptor ligand fulvestrant linked to cereblon as the E3 ligase. Effective degradation of the estrogen receptor has been demonstrated along with antitumor activity in breast cancer model systems.
PROTAC VZ185 degrades the bromodomain protein BRD9 and its close homolog BRD7. BRD9 is a component of the SWI/SNF (SWItch/Sucrose Non-Fermentable) chromatin remodeling complex, a group of proteins that remodel the way DNA is packaged. BRD9 has been implicated in various cancers as a regulator of tumor cell growth. The VZ185 PROTAC consists of a BRD9 ligand linked to an E3 ligase ligand.
The repertoire of targeted protein degradation has expanded in recent years to include new strategies to modulate protein levels. Molecular glues and monovalent degraders are alternative approaches to achieve targeted protein degradation (Fig. 4). Molecular glues are small molecules that bind to the target protein or E3 ligase, bringing them into proximity and promoting the degradation of the target protein. Molecular glues are not bifunctional like PROTACs but consist of a single molecule that induces interaction between the target protein and the E3 ligase. Besides monovalent degraders, there is a novel class of bivalent degraders that bind two different binding sites at the target of interest, thereby favouring the formation of a ternary complex. Read more on this in the application note: Studying intramolecular bivalent glues using TR-FRET binding assays.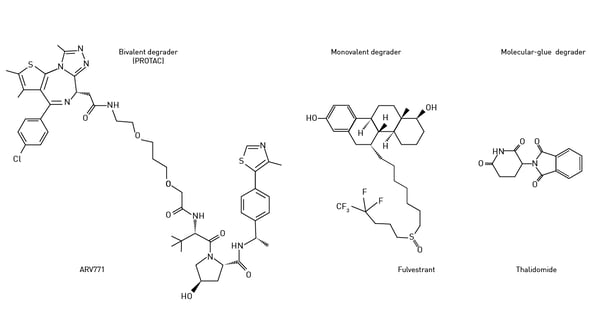 Stuart Schreiber coined the phrase “molecular glue” back in 1992 in the context of immunophilins.3 Cyclosporin and cyclophilin were amongst the first molecules shown to work in this way.
Stuart Schreiber coined the phrase “molecular glue” back in 1992 in the context of immunophilins.3 Cyclosporin and cyclophilin were amongst the first molecules shown to work in this way.
Examples of molecular glues that induce degradation of protein targets are thalidomide, lenalidomide, and pomalidomide (immunomodulatory or IMiDe drugs). Each of these degraders brings the target protein into proximity with cereblon, the E3 ubiquitin ligase mentioned earlier. Thalidomide, a potent teratogen, was initially developed as a sedative and anti-inflammatory agent but was later found to have anti-angiogenic and immunomodulatory effects. Thalidomide leads to protein degradation through its ability to bind the cereblon E3 ligase (Fig.5). 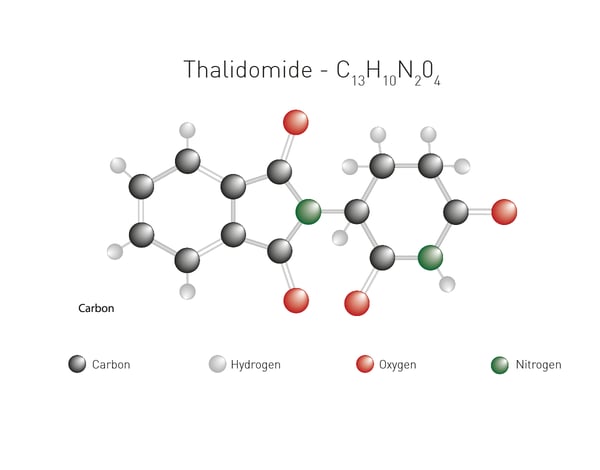 Monovalent degraders are designed with only one binding domain that specifically recognizes the target protein. The monovalent degrader binds to the target protein and recruits the endogenous E3 ligase machinery to induce protein degradation. This approach relies on the natural interaction between the target protein and the E3 ligase.
Monovalent degraders are designed with only one binding domain that specifically recognizes the target protein. The monovalent degrader binds to the target protein and recruits the endogenous E3 ligase machinery to induce protein degradation. This approach relies on the natural interaction between the target protein and the E3 ligase.
Microplate readers are versatile tools for PROTAC research. They are commonly used to measure different parameters and to assess the efficacy of PROTAC-mediated protein degradation. Examples of applications relevant to work with PROTACs include cell viability and proliferation assays, reporter gene assays and protein quantification.
Luminescence and other luminescense-derived technologies are powerful tools to look at protein-ligand binding for PROTACs. The application note Measuring protein ligand binding with an endogenous HiBiT CETSA test system describes luminescence assays that screen for PROTAC and protein of interest (POI) interactions (Fig. 6).
A NanoLuc® reporter enables the detection of low cellular protein levels by the HiBiT CETSA assay. The HiBiT CETSA assay combines the Cellular Thermal Shift Assay (CETSA), which permits measurement of endogenous full-length protein in a tracer-free manner, and HiBiT, a bioluminescent detection method based on protein complementation. The HiBiT tag is added to the protein of interest and detected when the complementary LgBiT subunit binds to HiBiT forming a functional luciferase. The HiBiT CETSA was successfully performed in 10 µl volumes using the PHERAstar® FSX. For more information on these kinds of experiments and more ways to evaluate PROTACs and other protein degrader check out this scientific talk:

NanoBRET™ and HiBiT enable binding and ubiquitination to be measured in live cells. This is further empowered by microplate readers equipped with temperature incubation and atmospheric control. Instruments like the CLARIOstar® Plus even allow long-term kinetic experiments to be run in the plate reader while keeping a physiological environment for cells.
In the application note Elucidating PROTAC MoA with live cell kinetic monitoring of ternary complex formation and target protein ubiquitination, a CLARIOstar with Atmospheric Control Unit was used to monitor PROTAC function in live cells. The study used luminescence as the detection mode and NanoBRET™ ternary complex and ubiquitination assays developed by Promega (Fig. 7). The ability to measure the live cell kinetics of ternary complex formation with an E3 ligase as well as the efficiency with which the target protein is ubiquitinated are essential for PROTAC optimization. This helped confirm the mode of action and to understand the efficacy of degradation.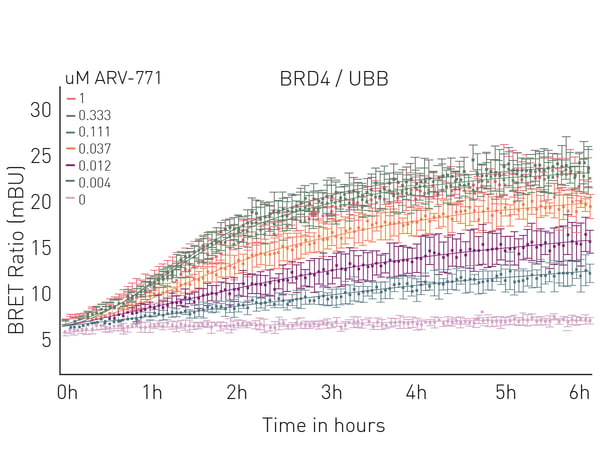 Fluorescence polarization assays can also be used to study the binding activity of PROTACs to their targets and the kinetics of interactions. Changes in fluorescence polarization due to fluorescently labeled PROTACs binding to target proteins can also be measured. This can provide information about the binding strength and kinetics of the interaction between the target protein and PROTAC.
Fluorescence polarization assays can also be used to study the binding activity of PROTACs to their targets and the kinetics of interactions. Changes in fluorescence polarization due to fluorescently labeled PROTACs binding to target proteins can also be measured. This can provide information about the binding strength and kinetics of the interaction between the target protein and PROTAC.
The application note Ubiquitination monitoring in real-time: the fluorescence polarization-based method UbiReal describes a fluorescence polarization method that can be used to track all stages of ubiquitin conjugation and deconjugation in real time. The approach is suitable for high throughput formats and fluorescence polarization is used to measure fluorescently labeled ubiquitin. Significantly, all stages of the ubiquitination cycle can be monitored using UbiReal (Fig. 8). PROTAC research has been held back by a paucity of tools to look at all stages of the protein ubiquitination cycle. UbiReal fills that gap.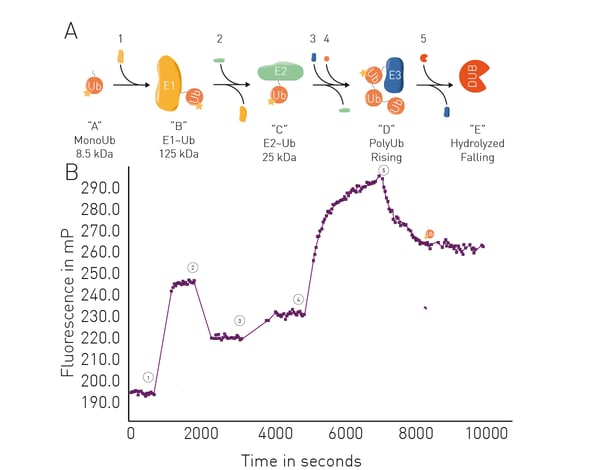 Microplate readers can also be used to assess protein-protein interactions involved in PROTAC-mediated protein degradation. For PROTACs, techniques like AlphaScreen® , Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) or luminescence methods like NanoBRET™ can be performed in microplates to measure the interaction between target and ligase.
Microplate readers can also be used to assess protein-protein interactions involved in PROTAC-mediated protein degradation. For PROTACs, techniques like AlphaScreen® , Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) or luminescence methods like NanoBRET™ can be performed in microplates to measure the interaction between target and ligase.
In addition, high-throughput screening techniques can be used to prioritize potential PROTACs and microplate readers are a useful tool that scales to the needs of the screen.
More than 15 targeted protein degraders have advanced to the clinic by the end of 2021.4 Most of the PROTACs in development use the cereblon E3 ligase and target cancer indications. Examples include ARV-10 and ARV-471 mentioned earlier for prostate and breast cancer, respectively, which are the most advanced candidates and are in phase II trials (as of 2022). Several molecular glues are also in clinical development. CC-220, CC-92480 and CC-90009 are in phase II trials for multiple myeloma and acute myeloid leukemia. Other areas of interest include chronic myeloid leukemia and solid tumors.
Researchers are looking at new ways to further advance the use and optimization of PROTACs as part of drug discovery efforts. To improve selectivity for the target protein and ligase, ligands are being refined through improved designs that strengthen affinity and specificity. Von Hippel-Lindau (VHL) and cereblon proteins have been the E3 ligases of choice in most PROTACs. However, there is a need to expand the ligase repertoire to target a wider range of proteins.
This topic is further discussed in “Developing next-generation therapeutics with targeted protein degradation”, an interview with Helen Harrison, Director of Screening at Amphista Therapeutics.
Most PROTACs target intracellular proteins but opportunities in drug discovery also exist to target extracellular and membrane-bound proteins. Inspired by PROTACs, researchers are developing protein degradation platforms that target degradation of extracellular and membrane-associated proteins using lysosome-targeting chimeras (LYTACS). Extracellular and membrane-associated proteins comprise 40% of all protein-encoding genes and are key agents in cancer, ageing-related diseases, and autoimmune disorders. 5
Tissue-specific targeting is another area of interest. The design of PROTACs with properties that enable selective accumulation and activity in specific tissues and organs could improve efficacy and minimize off-target effects. Combination therapies hold promise for synergistic effects between proteolysis targeting chimeras and other drugs and may help overcome mutations that lead to drug resistance.
New approaches like these should open exciting opportunities for future drug development efforts, preclinical testing, clinical trials and, ultimately, translation to the clinic.
What is the preferred BMG LABTECH microplate reader for specific needs and applications related to PROTAC research? BMG LABTECH offers a range of detection devices for sensitive absorbance, fluorescence and luminescence measurements.
The PHERAstar FSX was specifically conceived for screening campaigns and is your go-to reader for high-performance high-throughput investigations in PROTAC research.
Both the VANTAstar® and CLARIOstar Plus allow for wavelength scanning and include Enhanced Dynamic Range technology for superior performance in a single run. They also offer increased light transmission and sensitivity courtesy of Linear Variable Filter MonochromatorsTM and different filter options. In addition, they can be equipped with the Atmospheric Control Unit for live cell-based assays.
All BMG LABTECH microplate readers have exceptionally fast reading capabilities. In addition, the Omega series, CLARIOstar Plus and PHERAstar FSX microplate readers come with on-board injectors that can offer the very best options for detection at the time of injection.
Collectively, these multi-mode readers combine high performance with miniaturized assays, short measurement times, and offer considerable savings on materials and other resources.
Powerful and most sensitive HTS plate reader
Most flexible Plate Reader for Assay Development
Upgradeable single and multi-mode microplate reader series
Flexible microplate reader with simplified workflows
Degrons are specific sequences of amino acids or structural motifs within a protein that are important for targeted protein degradation. Find out how microplate readers can advance research into natural and engineered degrons.
Gene reporter assays are sensitive and specific tools to study the regulation of gene expression. Learn about the different options available, their uses, and the benefits of running these types of assays on microplate readers.
Innovation for targeted protein degradation and next-generation degraders is gathering pace. This blog introduces some of the different approaches that act via the lysosome or proteasome.
The choice of assay for targeted protein degradation studies is crucial. But what is the preferred assay and detection technology for your specific research needs and microplate reader?
Molecular glues are small molecules that help target unwanted proteins for destruction by the ubiquitin-proteasome system. Find out how microplate readers can advance molecular glue research.
Cannabinoids offer exciting opportunities to target diverse diseases with unmet needs. Learn how microplate readers can help improve our understanding of drug screening and drug signaling events to help advance cannabinoid research.