
PHERAstar FSX
Powerful and most sensitive HTS plate reader
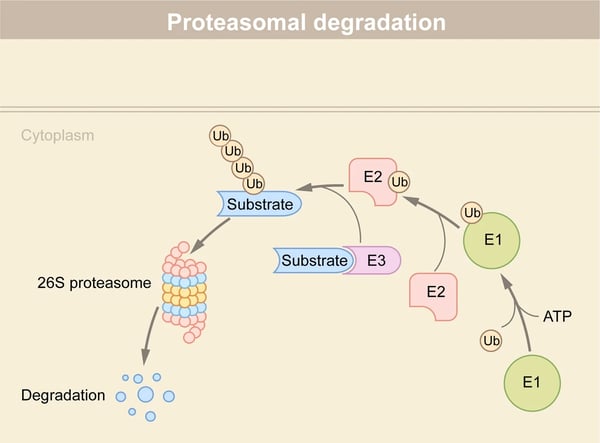
Molecular glues are small molecules that help target unwanted proteins for destruction in the cell’s ubiquitin-proteasome system. Find out how microplate readers can advance research into molecular glue degraders.
 Dr Barry Whyte
Dr Barry Whyte
Molecular glues offer exciting opportunities for targeted protein degradation and new ways to reach some of the estimated 85% of undruggable targets in the proteome. In this blog, we take stock of progress, look at what’s in store for drug development, and give examples of ways microplate readers can help advance research into molecular glues.
Molecular glues have been around for a while. Cyclosporin A (Fig.3) was first discovered in 1971 and approved by the FDA in 1983 as an immunosuppressant to prevent organ transplant rejection. However, it was not until 1991 that the mechanism of cyclosporin A as a molecular glue was revealed. In 1992, the phrase “molecular glue” finally found its way into a peer-reviewed publication describing how antigenic peptides selectively glue specific major histocompatibility complex (MHC) and T cell receptor proteins together to mount an immune response.3
 PROTACs arrived later on the scene and proof of concept for Protac-1, the first fully synthetic PROTAC, was reported in 2001.2 Since 2001, different technologies have proliferated expanding applications and opening new opportunities for clinical development (Fig.4).
PROTACs arrived later on the scene and proof of concept for Protac-1, the first fully synthetic PROTAC, was reported in 2001.2 Since 2001, different technologies have proliferated expanding applications and opening new opportunities for clinical development (Fig.4).
Examples of molecular glues that induce degradation of protein targets include thalidomide (Fig.5), lenalidomide, and pomalidomide (so-called immunomodulatory or IMiD drugs). Each of these degraders brings the target protein into proximity with cereblon, the most used E3 ubiquitin ligase to date.
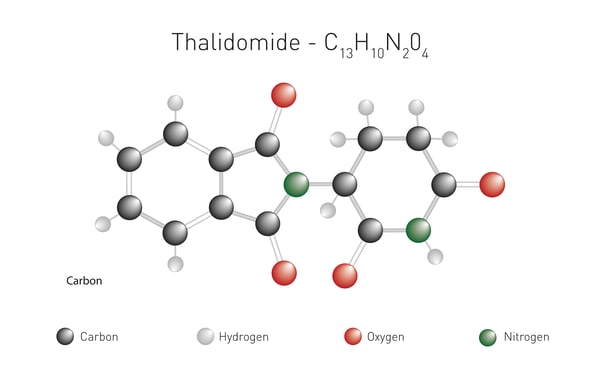 Thalidomide is perhaps one of the most well-known small drugs in the molecular glue arsenal. In the early 1960s, it was developed as a treatment for nausea in pregnant women but had to be withdrawn due to the serious birth defects it caused in children. Subsequently, thalidomide found other uses and was approved for the treatment of certain cases of leprosy in 1975. New derivatives of thalidomide followed. In 2006, lenalidomide (Revlimid) was approved by the FDA for the treatment of multiple myeloma. Eight years later it was shown that lenalidomide enabled the ubiquitination and degradation of hard-to-reach transcription factors. Further improvements to lenalidomide gave rise to pomalidomide, an anti-cancer medication used for the treatment of multiple myeloma and AIDS-related Kaposi sarcoma.
Thalidomide is perhaps one of the most well-known small drugs in the molecular glue arsenal. In the early 1960s, it was developed as a treatment for nausea in pregnant women but had to be withdrawn due to the serious birth defects it caused in children. Subsequently, thalidomide found other uses and was approved for the treatment of certain cases of leprosy in 1975. New derivatives of thalidomide followed. In 2006, lenalidomide (Revlimid) was approved by the FDA for the treatment of multiple myeloma. Eight years later it was shown that lenalidomide enabled the ubiquitination and degradation of hard-to-reach transcription factors. Further improvements to lenalidomide gave rise to pomalidomide, an anti-cancer medication used for the treatment of multiple myeloma and AIDS-related Kaposi sarcoma.
Additional molecular glues are in clinical development. CC-220, CC-92480 and CC-90009 are in phase II trials for different indications including systemic lupus erythematosus, multiple myeloma, and acute myeloid leukemia.
In parallel, interest in molecular glue degraders has gathered momentum as further scientific discoveries have expanded the research horizon. Molecular glues discovered so far have mostly been the products of chance. While the evolution of PROTACs has been driven by design principles for linkers and warheads, the molecular features of effective glues remain largely unknown. However, discoveries illuminating the way molecular glues work continue to bring new developments that make rational design more feasible.
Most molecular glues bind at one point of contact either to the ligase (most frequently) or the protein of interest. However, a new class of molecular glues, , so called intramolecular bivalent glues, was recently identified that involves a degrader attaching itself at two places to the target protein. This work used structural findings of the ternary complex of target protein, degrader and ligase to guide the rational design of improved degraders with low picomolar potency.
The study "Targeted protein degradation via intramolecular bivalent glue" was published in Nature by a scientific team that included researchers at the University of Dundee in the United Kingdom and the Research Center for Molecular Medicine (CeMM) in Austria.4
A crucial finding in the study was the identification of a structural rearrangement of the target protein due to the binding of the degrader at two points of contact. This rearrangement stabilized the interaction of the target protein with the E3 ligase and was crucial for the targeted protein degradation. Another example for a HiBiT based degradation assay can be found in the application note: dTAG protein degradation assay for the targeted degradation of proteins of interest.
In the study, HiBiT degradation assays were used to measure the potency of degradation and the maximum level of degradation possible for three different molecular glue degraders. The HiBiT degradation assays were carried out using the Nano-Glo® HiBiT Lytic Detection System from Promega and detection was performed on the PHERAstar® FSX. The degraders used in the study (IBGs 1-3) targeted the bromodomain-containing proteins BRD2 and BRD4 which are implicated in cancer and inflammation (Fig.6). The study opens a new area for exploration where protein degraders act by bridging protein domains in cis to enhance the fit with E3 ligases for productive ubiquitination and degradation. 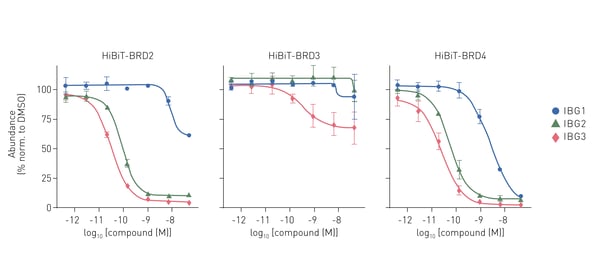
Rational design of molecular glues remains in its infancy but is clearly an area of interest for leading edge companies looking to combine artificial intelligence-guided methods with known databases of structures of molecular glues. Much interest lies in better understanding the role of degrons, elements on proteins that act as ubiquitin ligase recognition signals for protein degradation.
Many of the tags used today that work with molecular glues are too large to label the genes that encode a cell’s native proteins or act too widely beyond the targeted protein. To overcome this limitation, researchers at the Broad Institute of MIT and Harvard University developed a continuous evolution platform called PACE that generates smaller protein degradation tags (degrons) that form molecular glue complexes with the specificity needed to trigger depletion of the target protein. The compact degron is introduced into the cell’s genome by prime editing where it recruits the ubiquitin ligase cereblon.5 The PACE platform can be used to evolve molecular glue complexes with novel degrons.
A big search is also on for new ubiquitin ligases. Cereblon dominates the landscape for molecular glues discovered to date. However, beyond cereblon there are more than 600 known human ubiquitin E3 ligases. Some should offer specificity for certain cell types which would be an important step forward in limiting off target effects for future molecular glues.
This topic is further discussed in “Targeted protein degradation – next-generation therapeutics”, an interview with Helen Harrison, Director of Screening at Amphista Therapeutics.
Most molecular glue degraders target intracellular proteins, but opportunities exist to target extracellular and membrane-bound proteins. We will consider other emerging degrader technologies, including those that make use of the lysosomal degradation pathway, in more detail in a future blog.
Molecular glues have potential to unlock new therapeutic opportunities for a wide range of diseases including cancer, neurodegenerative disorders and autoimmune conditions.6 The potential for molecular glues as therapeutics is immediately relevant to many disease-related non-enzymatic proteins where traditional small molecule inhibitors that bind to an active site have proven to be ineffective. New approaches like these should open exciting opportunities for future small molecule drug discovery, drug development, high-throughput screening, preclinical testing, clinical trials and, ultimately, translation to the clinic.
Microplate readers are versatile tools for research into molecular glues and can be used to assess the efficacy of molecular glue-mediated protein degradation. Moreover, screening for molecular glue candidates typically requires high-throughput capabilities. Here microplate readers help increase throughput and decrease the need for manual intervention.
Luminescence and other luminescence-derived technologies are powerful ways to look at protein-ligand binding for molecular glues. NanoBRET™ and HiBiT enable binding and ubiquitination to be measured in live cells. This is further empowered by microplate readers equipped with temperature incubation and atmospheric control. Instruments like the CLARIOstar® Plus even allow long-term kinetic experiments to be run in the plate reader while keeping a physiological environment for cells. An example of using NanoBRET for ubiquitination assays on the CLARIOstar® Plus is shown in Fig.7 taken from the application note Elucidating PROTAC MoA with live cell kinetic monitoring of ternary complex formation and target protein ubiquitination.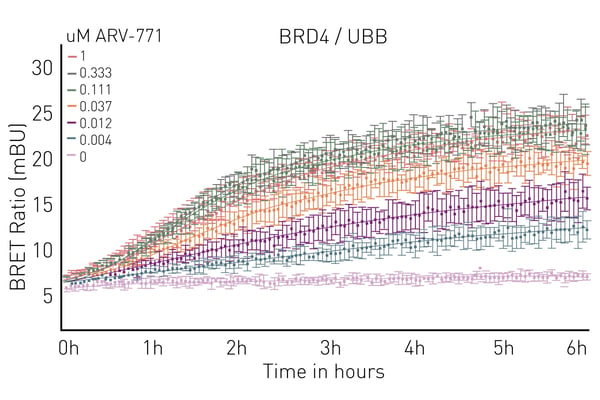
Fluorescence polarization assays can also be used to study the binding activity of molecular glues to their targets and the kinetics of interactions. Changes in fluorescence polarization due to fluorescently labeled molecular glues binding to target proteins may be measured. This can provide information about the binding strength and kinetics of the interaction between the target protein and degrader.
The application note Ubiquitination monitoring in real-time: the fluorescence polarization-based method UbiReal describes a fluorescence polarization method that can be used to track all stages of ubiquitin conjugation and deconjugation in real time. The approach is suitable for high throughput formats and fluorescence polarization is used to measure fluorescently labeled ubiquitin. All stages of the ubiquitination cycle can be monitored using UbiReal (Fig.8). 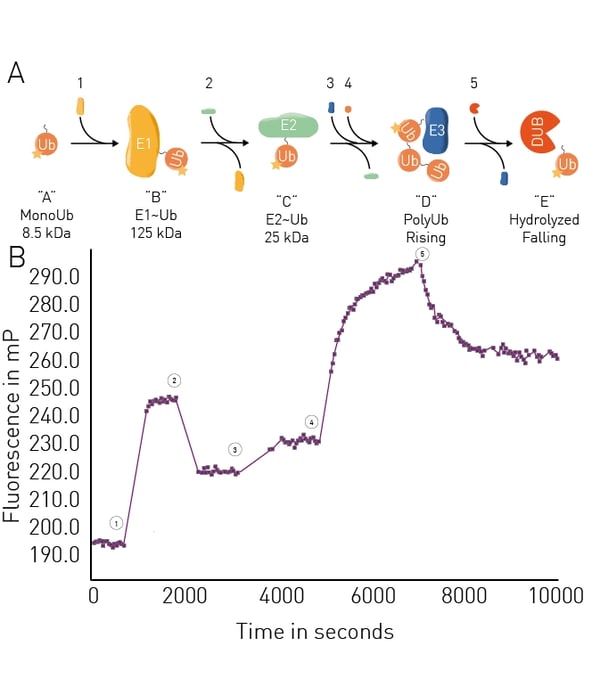
Microplate readers can also be used to assess protein-protein interactions involved in molecular glue-mediated protein degradation. Techniques like AlphaScreen®, Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) or luminescence methods like NanoBRET™ can be performed in microplates to measure the interaction between target and ligase.
In addition, high-throughput screening techniques can be used to prioritize potential molecular glue degraders and microplate readers are a useful tool that scales to the needs of the screen.
What is the preferred BMG LABTECH microplate reader for specific needs and applications related to molecular glue degrader research? BMG LABTECH offers a range of detection devices for sensitive absorbance, fluorescence and luminescence measurements.
The PHERAstar FSX was specifically conceived for screening campaigns and is your go-to reader for high-performance high-throughput investigations in molecular glue research.
Both the VANTAstar® and CLARIOstar Plus allow for wavelength scanning and include Enhanced Dynamic Range technology for superior performance in a single run. They also offer increased light transmission and sensitivity courtesy of Linear Variable Filter MonochromatorsTM and different filter options. In addition, they are recommended for live cell-based assays as they can be equipped with the Atmospheric Control Unit for CO2 and O2 regulation in the instrument.
All BMG LABTECH microplate readers have exceptionally fast reading capabilities. In addition, the Omega series, CLARIOstar Plus and PHERAstar FSX microplate readers come with on-board injectors that can offer the very best options for detection at the time of injection.
Collectively, these multi-mode readers combine high performance with miniaturized assays, short measurement times, and offer considerable savings on materials and other resources.
For more information on targeted protein degrader experiments and more ways to evaluate molecular glues, PROTACs and other degraders check out this scientific talk:

Powerful and most sensitive HTS plate reader
Most flexible Plate Reader for Assay Development
Upgradeable single and multi-mode microplate reader series
Flexible microplate reader with simplified workflows
Degrons are specific sequences of amino acids or structural motifs within a protein that are important for targeted protein degradation. Find out how microplate readers can advance research into natural and engineered degrons.
Gene reporter assays are sensitive and specific tools to study the regulation of gene expression. Learn about the different options available, their uses, and the benefits of running these types of assays on microplate readers.
Innovation for targeted protein degradation and next-generation degraders is gathering pace. This blog introduces some of the different approaches that act via the lysosome or proteasome.
The choice of assay for targeted protein degradation studies is crucial. But what is the preferred assay and detection technology for your specific research needs and microplate reader?
Cannabinoids offer exciting opportunities to target diverse diseases with unmet needs. Learn how microplate readers can help improve our understanding of drug screening and drug signaling events to help advance cannabinoid research.