
PHERAstar FSX
Powerful and most sensitive HTS plate reader
The investigation of binding kinetics is a crucial for drug research. Find out, how binding kinetics work and which approaches can be used to study them on microplate readers.
 Dr Martin Mangold
Dr Martin Mangold
Molecular interactions lie at the heart of biology. Binding kinetics experiments study the interaction behaviours of two or more biomolecules with each other over time and are the foundation of pharmacological theory. But what is the motivation behind performing such experiments?
Binding interactions are crucial events which rely on the proximity of two biomolecules to each other as well as on their affinity. Without binding interactions there would be no enzymatic activity, no receptor signalling, no cellular transport and no DNA transcription and translation. In short, life would not be possible without them!
Missregulated binding events, on the other side, can cause various diseases. These missregulations can express themselves for example in the accumulation of protein aggregates such as in Alzheimer´s and prion disease or in unwanted enzymatic activity which can lead to irregular degradation processes such as in autoimmune diseases. Here, studying binding kinetics is a viable tool for better understanding the underlaying mechanisms of a disease and identifying suitable targets for its treatment.
Accordingly, binding kinetics are an important aspect of drug research. Drugs are often specifically designed to target and eventually inhibit specific binding interactions. Binding kinetics are for example used in drug discovery studies for the screening of large libraries of small molecules for potential therapeutics. These drugs have to be as potent and specific as possible to prevent undesirable side effects. Kinetic parameters which can be determined in binding kinetics, such as binding affinity, are crucial tools for this purpose and provide information about different aspects of the interaction. Common targets for drugs comprise kinases and GPCRs which are the focus of many studies 1, 2, but also other protein-protein interactions regarding the proteasome or other proteases. Find out more about the importance of binding kinetics in the pharmaceutic industry in this scientific talk of Prof. Steven Charlton with the topic: The kinetics of drug binding - Why it is important?

To understand binding kinetics, it is important to first understand the principles behind binding interactions. Binding interactions are made possible by various different forces which allow inter-molecular connections such as Van der Waal´s forces, hydrogen bonding, electrostatic forces and hydrophobic effects. While most interactions are non-covalent and reversible, some binding interactions are based on covalent bonds that are formed between biomolecules. Watch this scientific talk of Martin Redhead, group leader at UCB, to find out more about the advantages of kinetic binding methods in contrast to binding affinity analysis at equilibirium.

Independent of the type of binding interaction, it is important to note, that binding is not static. The interaction of two biomolecules is comprised of association events according to their association rate and dissociation events according to their dissociation rate. Binding changes over time until it reaches an equilibrium at which the concentrations of binding complexes are constant. Accordingly, binding experiments are generally performed over time as binding kinetics. Binding kinetics can also be used to analyse other factors that can influence the time course of an interaction such as for example temperature, pH, viscosity, or the concentrations of interaction partners.
In binding kinetics three parameters are particularly important. The association, or on-rate kon, defines how much of a ligand (L) binds to a biomolecule (B) over a specific time to form the biomolecule ligand complex (BL). The dissociation, or off-rate koff, on the other hand defines the amount of ligand that dissociates from the biomolecule ligand complex over a specific time. Together they form the equilibrium dissociation constant KD of a ligand which is the ratio of dissociation over association events over time. KD indicates the affinity of a ligand to a biomolecule and is an important parameter for binding and drug research: The lower the KD value the higher the affinity of a ligand or drug to its target which means lower ligand concentrations are needed to induce binding. However, since high affinity can be the cause of either a low koff rate or a high kon rate it is important to determine these parameters in binding kinetics to fully understand a drug’s or ligand’s binding behaviour and choose the right administration. For example, drugs with a high kon rate have a faster onset of action, while the effect of drugs with a low koff rates lasts longer since the drug remains bound to its target for a longer time.
Dependent on the investigated biomolecules, desired readout and kinetic parameters, different approaches can be followed to study binding kinetics.
Now that the significance of binding kinetics has been clarified, the question naturally arises as to how the study of binding kinetics can be carried out? Below I will focus on the four approaches which are used most often to study binding kinetics on microplate readers.
Förster´s Resonance Energy Transfer (FRET) and time-resolved FRET (TR-FRET) are probably the most popular detection modes used for performing binding kinetics. Both are based on the energy transfer between two fluorophores. These fluorophores are attached to the two binding partners of a binding interaction. Since they have overlapping emission and excitation spectra, emission of the donor fluorophore leads to the subsequent excitation of the acceptor fluorophore, but only when both fluorophores and binding partners are in close proximity. However, since FRET can be negatively affected by scattered excitation light and autofluorescence in a sample, it can suffer from high background signals. To eliminate this problem, TR-FRET was developed, which uses long-lived lanthanide donor fluorophores to reduce background signals.
The application note "Binding kinetics: high throughput assay for kinase inhibitors" from BMG LABTECH´s application note library highlights a microplate-based TR-FRET assay for performing binding kinetics. Here, the assay was used to determine the target binding kinetic rates kon and koff for kinase inhibitors. This was achieved by sequentially performing association and dissociation binding kinetics (fig. 1) and allowed screening of 270 potential inhibitors against a panel of 40 kinase drug targets in a high-throughput manner. The versatility of TR-FRET assays for performing binding kinetics is highlighted in further application note examples, such as the analysis of binding kinetics with HTRF or the detection of human tau protein aggregation.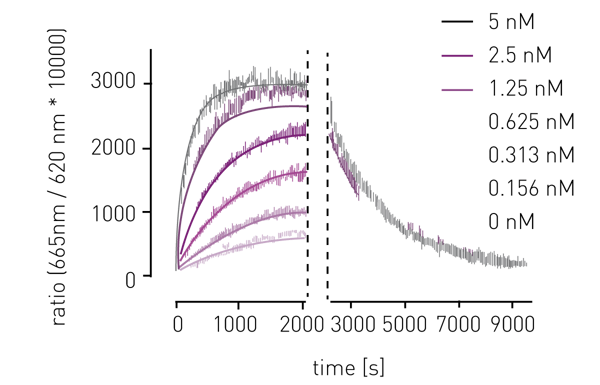
BRET assays are also based on the resonance energy transfer between two interaction partners and can thus be used to perform binding kinetics. In contrast to FRET, the donor signal in BRET assays is provided by a luciferase which generates emission light by an enzymatic reaction (fig. 2). Accordingly, no additional excitation source has to be defined. As with FRET, BRET assays are performed as ratiometric measurements of acceptor and donor signal offering comparability between different test runs.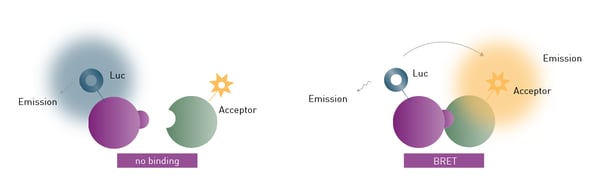 However, the available BRET assay systems BRET1 and BRET2 have some shortcomings regarding sensitivity and background noise. To circumvent these, NanoBRET was developed, which offers greatly enhanced luminescence signal intensity and stability using a modified luciferase derived from a deep-sea shrimp species, which provides a emission signal 100x brighter than conventional firefly or renilla luciferases.
However, the available BRET assay systems BRET1 and BRET2 have some shortcomings regarding sensitivity and background noise. To circumvent these, NanoBRET was developed, which offers greatly enhanced luminescence signal intensity and stability using a modified luciferase derived from a deep-sea shrimp species, which provides a emission signal 100x brighter than conventional firefly or renilla luciferases.
NanoBRET was employed in the application note "NanoBRET™ assay quantitatively evaluates VEGF binding to the VEGFR2 in real-time in living cells" to study the binding kinetics of vascular endothelilal growth factor (VEGF) to its receptor VEGFR2 (fig. 3). Here, a novel labelling technique was used to label multiple VEGF isoforms and quantitatively evaluate their binding to VEGFR2 in real-time in living cells. In another approach, the same was achieved for the monitoring of ligand binding to GPCRs. Alternatively, BRET assays can also be used to study G-protein dissociation as well as β-arrestin recruitment to GPCRs upon ligand binding. Novel luminescence-based approaches use the NanoBIT® system, which employs a fragmented nanoluciferase attached to two binding partners of interest. Upon binding of the two binding partners, the nanoluciferase fragments form a functional enzyme that can generate a luminescence signal as highlighted in the application note: Identification of androgen-disruptors using a cell-based androgen receptor dimerization assay.
Fluorescence polarization assays use polarized excitation light and measure the polarization of emission light to investigate a biomolecule and its binding kinetics. It is most often used to study protein-protein and protein-DNA binding, as well as enzyme activity.
Fluorescence polarization uses small fluorescent ligands which on their own have a rapid tumbling rate. Accordingly, when the polarized excitation light excites the fluorescence ligand, the emission light becomes depolarized. After binding to a larger biomolecule, the tumbling rate of the fluorescent ligand is reduced, and its emission light retains most of its polarization (fig. 4). Comparing the polarization of unbound to bound fluorescent ligand allows to perform binding kinetics.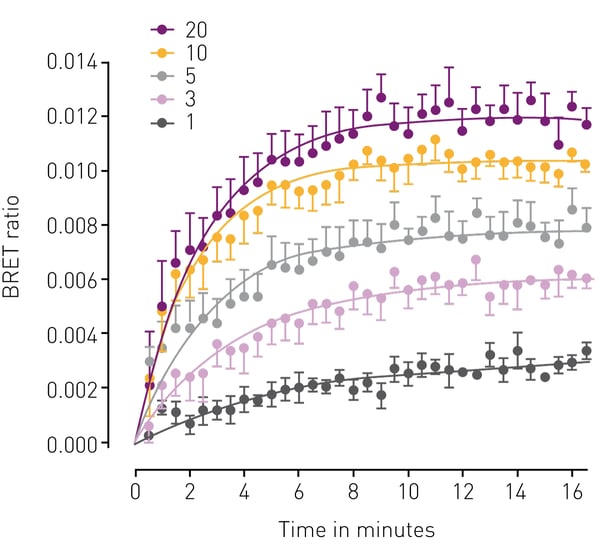 This principle was used in the application note "Protein-ligand binding measurements using fluorescence polarization" to investigate the binding kinetics of human Lys-specific histone demethylase 1 (LSD1) to histone derived peptides. Here, binding assays for three different peptides were performed, followed by competition experiments with potential competitors. This Application Note on a Competition assay using Fluorescence Polarisation to determine the Residence Times for Calcitonin and AMYR agonist, AM833, demonstrates, that fluorescence polarization can also be employed to determine association and dissociation rates of ligand receptor interactions. Fluorescence polarization is also suited for performing binding kinetics in high-throughput as highlighted in the application note "Fluorescence polarization based assay suitable for screening H-Prostaglandin D Synthase inhibitors", where this detection mode was used to screen for prostaglandin D synthase inhibitors.
This principle was used in the application note "Protein-ligand binding measurements using fluorescence polarization" to investigate the binding kinetics of human Lys-specific histone demethylase 1 (LSD1) to histone derived peptides. Here, binding assays for three different peptides were performed, followed by competition experiments with potential competitors. This Application Note on a Competition assay using Fluorescence Polarisation to determine the Residence Times for Calcitonin and AMYR agonist, AM833, demonstrates, that fluorescence polarization can also be employed to determine association and dissociation rates of ligand receptor interactions. Fluorescence polarization is also suited for performing binding kinetics in high-throughput as highlighted in the application note "Fluorescence polarization based assay suitable for screening H-Prostaglandin D Synthase inhibitors", where this detection mode was used to screen for prostaglandin D synthase inhibitors.
AlphaScreen® is a bead-based chemistry used to perform binding kinetics. Today AlphaScreen technology includes multiple assay formats, which are mainly used in the life sciences for screening purposes.
AlphaScreen uses specific donor and acceptor beads which are bound to the two interaction partners of interest, respectively. When in proximity energy transfer from one bead to the other leads to the generation of a luminescent light signal which can be quantified (fig. 5). The special feature of this method is that the excitation light has a longer wavelength than the measured emission signal.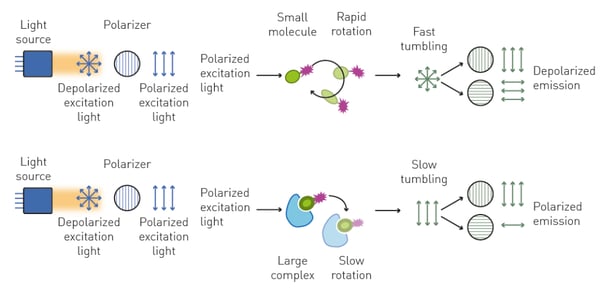 AlphaScreen was employed in the application note "PHERAstar measures AlphaScreen assay to develop selective inhibitors for the human YEATS domains" which highlights an assay that can be used for inhibitor screening. Here, several compound libraries with a total of over forty thousand potential inhibitors were screened against the ENL YEATS domain, yielding a potent and selective small molecule inhibitor (fig. 6).
AlphaScreen was employed in the application note "PHERAstar measures AlphaScreen assay to develop selective inhibitors for the human YEATS domains" which highlights an assay that can be used for inhibitor screening. Here, several compound libraries with a total of over forty thousand potential inhibitors were screened against the ENL YEATS domain, yielding a potent and selective small molecule inhibitor (fig. 6).
Even though binding kinetics can be performed by means of surface plasmon resonance or biolayer interferometry, microplate readers have established themselves as the gold standard due to several advantages. Because of the microplate format, binding kinetics could be substantially miniaturised, allowing the screening of large compound libraries in high throughput, minimising the needed measurement time as well as the cost for reagents and personnel.
But which of BMG LABTECH’s microplate readers are suited best for performing your binding kinetics experiments? For the multitude of available binding assays, BMG LABTECH multi-mode readers which can be equipped with more than one of the above-mentioned detection modes, such as the CLARIOstar® Plus or the VANTAstar®, offer the ideal solution between flexibility and sensitivity for all of your applications.
However, if you are looking for the most sensitive and fastest detection of binding kinetics possible, the PHERAstar® FSX is your go-to reader. It’s equipped with special features for this purpose, such as dedicated TRF and Alpha Technology lasers to increase TR-FRET and AlphaScreen assay performance. Furthermore, due to four dedicated PMT detectors, the PHERAstar can measure at two distinct emission wavelengths simultaneously with the Simultaneous Dual Emission (SDE) feature. Since most binding kinetics are performed as ratiometric measurements, this essentially reduces the necessary measurement time by half. At the same time, data quality is increased, since the two signals derive from the same excitation event and are less susceptible to other potential influencing variations.
Moreover, the AAS system provides a steady temperature that is unaffected by external environmental changes while the plate reader is operating. This provides improved assay stability for temperature-sensitive reactions such as binding reactions.
In his testimonial, Pioneering Pre-Clinical Drug Discovery with Advanced Assays, Nick Holliday from Excellerate Bioscience and the University of Nottingham discusses the benefits of the PHERAstar FSX for TR-FRET-based kinetic binding analysis.

Powerful and most sensitive HTS plate reader
Most flexible Plate Reader for Assay Development
Flexible microplate reader with simplified workflows
Degrons are specific sequences of amino acids or structural motifs within a protein that are important for targeted protein degradation. Find out how microplate readers can advance research into natural and engineered degrons.
Gene reporter assays are sensitive and specific tools to study the regulation of gene expression. Learn about the different options available, their uses, and the benefits of running these types of assays on microplate readers.
Innovation for targeted protein degradation and next-generation degraders is gathering pace. This blog introduces some of the different approaches that act via the lysosome or proteasome.
The choice of assay for targeted protein degradation studies is crucial. But what is the preferred assay and detection technology for your specific research needs and microplate reader?
Molecular glues are small molecules that help target unwanted proteins for destruction by the ubiquitin-proteasome system. Find out how microplate readers can advance molecular glue research.
Cannabinoids offer exciting opportunities to target diverse diseases with unmet needs. Learn how microplate readers can help improve our understanding of drug screening and drug signaling events to help advance cannabinoid research.