
SPECTROstar Nano
Absorbance plate reader with cuvette port
Antimicrobial resistance is a formidable problem across the world. Find out how microplate readers can help tackle resistant microbes.
 Dr Barry Whyte
Dr Barry Whyte
Antimicrobial resistance (AMR) is one of the top ten threats to public health worldwide.1,2
In a study published in The Lancet in 2022, over 1 million people were estimated to have died from bacterial AMR in 2019.3 The burden due to bacterial AMR is just the tip of the iceberg. Societal and scientific challenges span drug resistance across the microbial world.
In this blog, we look at research that offers solutions to tackle drug-resistant microbes. From small molecule inhibitors and ways to combat biofilms to emerging therapies like the use of bacteriophage. How can research help combat AMR in bacteria, fungi, and viruses and how can the use of microplate readers help scientists advance work to tackle resistant microbes?
The microbial world is incredibly diverse and far reaching. Not only do microbes inhabit water, soil, and the air but millions of them find a home in the human body too. The most common types of microbes in these environments are bacteria, viruses, and fungi. Some cause illnesses but others are important for human health.
Over the years, science has found ways to tackle some of the microbes that cause disease. Antibiotics have been a notable resource in the disease-fighting toolbox but increasingly AMR threatens to undo progress. Natural selection is at the heart of resistance to antibiotics in the microbial population. Drug-resistant microbes readily arise from errors in DNA replication and grow and expand through bacterial populations. At the same time, genes for resistance can mobilize and transfer themselves to different genomes and species. 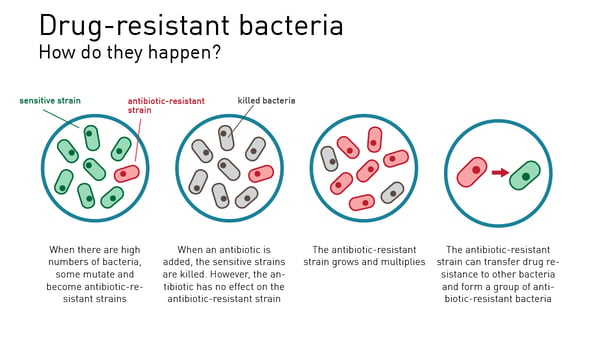
Solutions are needed at different levels due to the underlying biological complexity supporting drug resistance.3 For example, bacterial microbes have an intrinsic physical barrier (cell envelope) that offers ways to resist drug uptake. Bacterial microbes can also form biofilms on many types of surfaces that make drug entry even trickier. Once inside the bacterial cell, an antibiotic can face a barrage of intracellular challenges before ever reaching its molecular target.
Researchers not only need to find new drugs for molecular targets. They also need to find solutions that overcome the physical barriers and adaptive strategies adopted by microbes. It may take 8-20 years to develop and launch a new antibiotic via a traditional drug discovery effort.11 While this route provides some value, drug resistance works at breakneck speed.
The future will most likely not exclusively depend on the discovery of highly successful broad-spectrum drugs but will involve a “community” of approaches to treat and prevent infection.
Microplate readers are invaluable tools in antimicrobial research, including the search for new antimicrobials and the quest to improve existing treatment options. They are ideally suited to study microbial growth, assess antimicrobial activity, perform mode-of-action studies, and to screen at scale for potential antimicrobial compounds. They also allow for testing of antimicrobials on recalcitrant cell-based biofilms.
One of the most frequently encountered uses of microplate readers for microbes is the measurement of microbial growth. This can readily be done by measuring the optical density at 600 nm or OD600 for short. The ins and outs of measuring microbial growth are explained in detail in the BMG LABTECH blog Measure microbial growth using OD600. Here we provide some examples of specific applications relevant to antimicrobial resistance:
The application note Growth of Neisseria meningitidis in a BMG LABTECH microplate reader with Atmospheric Control Unit (ACU) demonstrates how N. meningitidis can be grown and studied under optimal growth conditions. Invasive meningococcal disease can result in a human fatality rate of 10% and N. meningitidis is becoming increasingly resistant to treatment with frontline antibiotics like penicillin, rifampicin, cefotaxime and ciprofloxacin.4 It is important to have ways to study the growth of N. meningitidis and to assess possible interventions.
The study showed that the proliferation of this organism was possible on a BMG LABTECH microplate reader supporting the 5% CO2 conditions required for optimal growth (Fig. 2). The use of a microplate reader and Atmospheric Control Unit is an ideal system for researchers looking to investigate the microbial growth of this organism under different conditions.
In the application note Growth of Campylobacter using a microplate reader equipped with ACU bacterial isolates were grown on microplates and growth assessed by measurements on a BMG LABTECH FLUOstar Omega reader equipped with an Atmospheric Control Unit. Antibiotic-resistant Campylobacter can spread from animals to humans and may contaminate the food chain. It is important to have ways to study these organisms as they grow. C. jeuni has strict requirements for growth. It will only grow in a defined temperature range and requires oxygen at a reduced level of 5-8% and elevated levels of CO2 of 10%. This study demonstrated that the growth of this fastidious organism could be achieved on a 96-well microplate and its proliferation effectively monitored by measuring the absorbance at 600 nm (Fig. 3).
Nephelometry is an alternative method to measure microbial growth. The NEPHELOstar Plus, a laser-based microplate nephelometer, can be readily used to make light scattering measurements on a microplate with low sample volumes at high sensitivity.
The application note Monitoring of microbial growth curves by laser nephelometry demonstrates the reliability of this technology for monitoring microbial growth. Corynebacterium glutamicum was used and growth curves were recorded on the NEPHELOstar Plus by measuring the turbidity of cultures at different time points.
One key advantage of using nephelometry is its ability to detect scattered light at low amounts of particles. This makes it particularly suited to the measurements required during the lag phase of microbial growth or at the beginning of the log phase since it offers higher accuracy and sensitivity.
In the application note Nephelometric monitoring growth of Candida albicans using BMG LABTECH’s NEPHELOstar® Plus a way to measure the propagation of a fungal microbe is described as well as the impact of antifungal agents on its growth (Fig. 4). In addition to monitoring the growth of C. albicans in the presence of antifungal agents, laser nephelometry can provide useful information on drug solubility.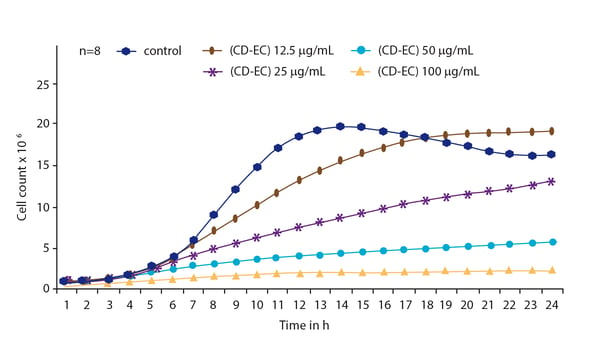
The minimal inhibitory concentration (MIC) is the lowest concentration of an antimicrobial compound or molecule that inhibits microbial growth. Microplate readers can be used to determine the MIC by measuring the light signal (absorbance, fluorescence or luminescence) of samples in a microplate containing different concentrations of an antimicrobial compound. The ability of a microplate reader to measure thousands of samples in a matter of minutes allows for quick and precise determination of the lowest concentration of inhibitor needed. This information is crucial for designing further experiments including high-throughput screening.
In some cases, it makes sense to evaluate the synergistic effects of different combinations of antimicrobials or antimicrobials and other molecules including adjuvants. This type of study may enhance effectiveness and help reduce the risk of resistance to treatment. Microplate readers can be used to test the different permutations quickly and at scale.
Biofilms are layers of microbial organisms that have aggregated to form a colony. The colony adheres to a surface and may be coated with a layer of polysaccharide (slime). Bacterial biofilms are particularly resistant to treatment with antibiotics. Microplate readers can be used to measure the ability of antibiotics to disrupt or prevent biofilm formation. They can also quickly identify the deficiencies of a particular intervention in breaching biofilm arrangements of a particular microbe (see Emerging areas of antimicrobial research in this blog, section on nanoparticles, for an example of a BMG LABTECH application).
Microplate readers can be used by researchers to simultaneously test many potential antimicrobials or combinations of antimicrobials in a rapid and automated manner. This type of high-throughput approach accelerates the discovery process by screening numerous compounds in parallel. Large libraries of chemical compounds can be tested to identify molecules with antimicrobial properties. Compounds that inhibit microbial growth are candidates for further investigation.
In the paper “High-throughput screening for bacterial glycosyltransferase inhibitors” a team of researchers performed a primary screen of a compound library looking for small molecule inhibitors that might be suitable for further development as alternatives to antibiotics.5 The authors used UDP-Glo luminescence assays on a FLUOstar Omega microplate reader to monitor UDP liberation. UDP is produced when the substrate UDP-GlcNAc is consumed by members of the glycosyltransferase NleB/SseK family in the presence or absence of the test compounds.
The NleB/SseK family are type III secretion system proteins that are appealing drug targets within enteropathogenic and enterohemorrhagic pathogens like E. coli and Salmonella enterica, respectively. Two compounds were identified as inhibitors suitable for further studies and possible refinement. This type of drug could potentially be used to replace or augment traditional antibiotic regimens for these pathogens.
The paper “A high-throughput screening strategy to identify inhibitors of SSB protein-protein interactions in an academic screening facility” describes how nine potent and selective inhibitors were identified in a pilot screen of 72,474 compounds for this target.7 Bacterial single-stranded DNA-binding protein (SSB) is a potential antibiotic target that serves as a hub for DNA repair, recombination, and replication. Mutations that disrupt protein-protein interactions within the C-terminal tail of SSB are lethal which suggests that inhibitors of these critical interactions could be effective antibacterial agents. Primary and secondary screening assays involved Alphascreen® and fluorescence polarization assays, respectively, performed in 1536-well microplates on a BMG LABTECH PHERAstar FSX microplate reader.
Shigella flexneri is a gram-negative bacterium that causes shigellosis, the most infectious of bacterial dysenteries. Shigellosis causes around 200 000 deaths a year.8 Shigella spp. that are resistant to almost all classes of antimicrobial drugs are increasing in prevalence across the world.
In the paper “Potential novel antibiotics from HTS targeting the virulence-regulating transcription factor, VirF, from Shigella flexneri”, researchers identified three small molecules that could block the cell-to-cell spread of this highly virulent pathogen from a screen of 140,000 molecules.9 The high-throughput beta-galactosidase reporter assays were measured on a BMG LABTECH PHERAstar FSX by looking at the absorbance of chlorophenol red at 570 nm. Bacterial growth was measured by monitoring the absorbance at 600 nm and the effects of inhibitors on growth determined. One of the identified compounds was also able to lower the initial invasion efficiency of S. flexneri which made it a possible candidate for hit-to-lead development.
New approaches are urgently needed to combat global AMR. Several innovations hold promise for the future of antimicrobial research.4
Conventional antibiotics are in many cases microbial natural products derived from a narrow window of related bacterial organisms (mainly the actinomycetes). One approach that holds promise for the future is to explore other genera for new chemical diversity. Previously uncultured bacteria are one untapped source for the discovery of next-generation antibiotics. Most bacterial diversity remains uncultured.10 New technologies have been developed to help this type of approach along. The iChip (isolation chip) is one example.11 The iChip consists of several hundred miniature diffusion chambers each inoculated with a single environmental cell. The chip is designed so that microbial recovery significantly exceeds traditional cultivation and ensures that the grown species are of significant phylogenetic novelty. Synthetic small molecules based on a better understanding of the physical chemistry of antimicrobials offer another avenue of exploration. Researchers are also looking to develop peptide-based antibiotics that are safe and have high efficacy.
A healthy gut can fight bacterial infections. Antibiotics not only target specific bacteria but can have an oversize effect on the friendly bacteria living in the gut. If this happens, it makes it easier for pathogenic bacteria to take hold. Researchers are looking at different ways to restore the balance of the microbiota to prevent bacteria like Clostridioides difficile from wreaking havoc after treatment with antibiotics. This work is at an early stage but represents one of several probiotic strategies attracting attention.
Nanoparticles can bypass drug-resistant mechanisms in bacteria and inhibit biofilm formation. These small particles, which have a diameter in the 1-100 nm range, penetrate the cell wall and membrane of bacteria and lead to bactericidal activities. Combined with antibiotic treatment, nanoparticles may also show synergy of action that could limit the amount of antibiotic needed for effective control of a pathogen and help relieve AMR. The precise mechanism of action of nanoparticles is in most cases unknown. Further research is needed to define exactly how they work and to find the optimal conditions for their possible clinical application.
The application note Detection of plant-synthesized nanoparticles and their antibacterial capacity describes the use of a microplate reader from BMG LABTECH to investigate the antibacterial properties of silver and zinc oxide nanoparticles on Vibrio cholerae and Escherichia coli. The amounts of the nanoparticles were determined, and cell growth of the bacteria was tracked during exposure to different levels of each nanoparticle. The antibacterial activity of the nanoparticles could be measured in a biofilm assay performed in 96-well microplates. The study demonstrated that silver and zinc oxide nanoparticles can be used to combat the formation of bacterial biofilms and is an effective method to help treat drinking water before it is consumed (Fig. 5).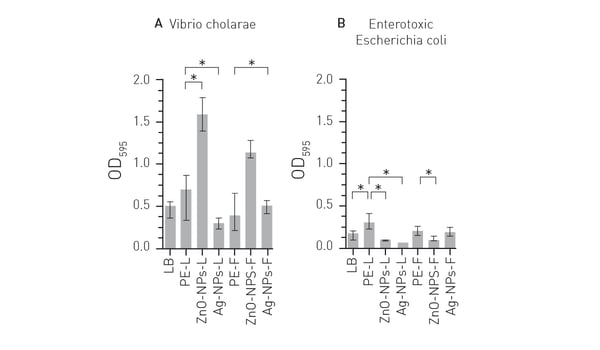
As highlighted by the Covid-19 pandemic, the prevention of infection is a preferred strategy in the control of disease. Vaccines against antimicrobial-resistant pathogens can therefore contribute significantly to fighting AMR by tackling the problem at source.4 The use of an effective vaccine against a specific pathogen translates into a need for less use of antibiotics and lower AMR. The discovery and development of mRNA vaccines has greatly reduced the time needed to introduce a safe and effective vaccine. An approach that involves integration of novel vaccine technology with new antibiotics, diagnostics, protein therapeutics, probiotic interventions and the use of bacteriophage may be required to combat AMR effectively.12
Quorum sensing is a type of cell-to-cell communication in bacteria that depends on secreted chemical signaling molecules called autoinducers. The process allows a whole population of bacteria to collectively regulate gene expression and therefore their behavior. Quorum sensing has been shown to regulate different processes in bacteria. These include bioluminescence, symbiosis, expression of virulence genes and surface proteins, as well as the formation of biofilms in Escherichia coli. The inhibition of quorum sensing can be used to suppress bacterial virulence since it places less selective pressure on pathogens and may avoid the development of resistant bacteria. As such, it represents an alternative approach to the use of antibiotics to fight bacterial disease.
In the application note Monitoring bacterial cell-to-cell communication quorum sensing using a BMG LABTECH microplate reader the effects of quorum sensing were investigated by measuring bioluminescence and bacterial growth in parallel. Different strains of Vibrio fischeri were grown under different defined environmental conditions to determine the impact of quorum sensing on this Gram-negative bacterium.
Bacteriophage are viruses that infect and replicate only in bacterial cells where they can cause cell lysis. As an intervention against bacteria, they offer high specificity, the possibility to administer many phage in a small dose, speed of development (months), and low toxicity to humans. Phage can also be used to deliver CRISPR-Cas therapeutics into bacteria. Researchers are looking at ways to bring bacteriophage treatments to the clinic. In 2015, no clinical trials for bacteriophage were on record. The number reached 71 in 2023.13
In the application note Testing novel bacteriophages for antibacterial properties with a crystal violet biofilm quantification assay a new bacteriophage was identified with antibacterial activity against four strains of antibiotic-resistant Escherichia coli. Bacterial biofilms were grown on the bottom of 96-well microplates and the extent of biofilm that remained after treatment with phage was assessed by crystal violet staining (Fig. 6). The newly identified bacteriophage had a wide range of lytic activity and broad stability making it an attractive candidate for further preclinical and clinical development.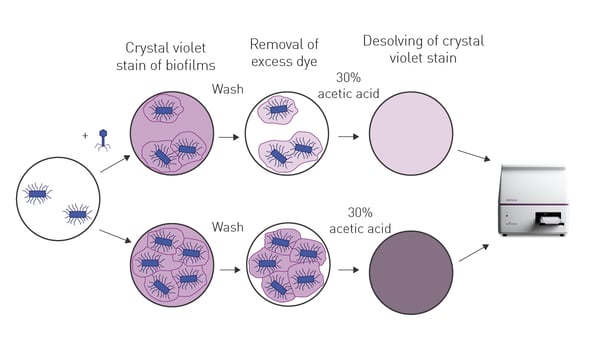 New approaches like these should open exciting opportunities for future research that complement existing strategies for AMR.
New approaches like these should open exciting opportunities for future research that complement existing strategies for AMR.
What is the preferred BMG LABTECH microplate reader for specific needs and applications related to antimicrobial resistance? BMG LABTECH offers a range of detection devices for sensitive absorbance, fluorescence and luminescence measurements.
All BMG LABTECH microplate readers have exceptionally fast reading capabilities. Collectively, these multi-mode readers combine high performance with miniaturized assays, short measurement times, and offer considerable savings on materials and other resources.
Absorbance plate reader with cuvette port
Powerful and most sensitive HTS plate reader
Most flexible Plate Reader for Assay Development
Upgradeable single and multi-mode microplate reader series
Flexible microplate reader with simplified workflows
Scientists measure bacterial growth in laboratories across the world but why is it so universal?
Quorum sensing is a type of cell-to-cell communication in bacteria that depends on cell density. Find out how microplate readers can help advance quorum sensing research.
You want to make sure that your cell culture is free of mycoplasma? BMG LABTECH microplate readers are the perfect measurement platform to read MycoAlert® mycoplasma detection kits.
OD600 is part of a lot of microbiology studies. Reason enough for us to use this article to explain why the method is used so intensively and what should be considered for the measurement.
Obligate anaerobic gut bacteria require an absolute anaerobic environment. Read how the FLUOstar Omega was used to develop new methods to study the human gut microbiome.