Introduction
The quantification of DNA is a standard laboratory activity and is pivotal for subsequent applications such as next generation sequencing (NGS). In addition to UV-spectrometry quantitation and qualification of nucleic acids, fluorescent probes provide the required specificity and precision to measure double-stranded DNA (dsDNA) during NGS sample preparation. The widely used Qubit™ nucleic acid staining dyes can be analyzed on a Qubit™ fluorometer. It measures fluorescence intensity directly in sample preparation tubes and automatically calculates the DNA-concentration based on two standards measured in parallel. This provides an easy, quick and intuitive way to quantitate dsDNA. However, throughput is limited using the Qubit™ fluorometer as only one sample can be analyzed at a time. This application note presents how the Qubit® dsDNA HS fluorophore can be used to quantitate dsDNA on a BMG LABTECH microplate reader.
Materials & Methods
- Qubit™ dsDNA HS Assay Kit (ThermoFisherScientific, #Q32854)
- TRIS-EDTA buffer 100x (Carl Roth GmbH & Co. KG)
- 96-well microplate (black, flat bottom, Greiner)
The Qubit working solution was prepared by mixing 60 µL of Qubit dsDNA HS reagent with 11.94 ml Qubit dsDNA buffer to obtain 12 ml working solution. Dilutions of the standard were prepared in 1x TE-buffer.
For Qubit DNA quantification in 96 well plates, 190 µl of Qubit working solution were mixed with 10 µl of DNA standards (0 and 10 ng/µl provided with the Qubit kit) or diluted DNA standard. Fluorescence intensity was measured on FLUOstar Omega, CLARIOstar, and PHERAstar with the following settings.
Instrument settings
|
Fluorescence intensity, endpoint protocol
|
|||
|
Optic settings
|
PHERAstar
|
Optic Modules
|
FI 485 520
|
|
CLARIOstar
|
Monochromator
|
Excitation: 483-14
Dichroic: auto 502.5
Emission: 530-30
|
|
|
Filters
|
Excitation Ex485
Dichroic: LP504
Emission: 530-40
|
||
|
FLUOstar Omega
|
Filters
|
Excitation: Ex485
Emission: Em520
|
|
|
Gain and focus adjusted prior to measurement
|
|||
|
Top optic
|
|||
|
General settings
|
Settling time: 0.2 s
|
0.5 s
|
|
|
Number of flashes per well
|
20
|
||
Results & Discussion
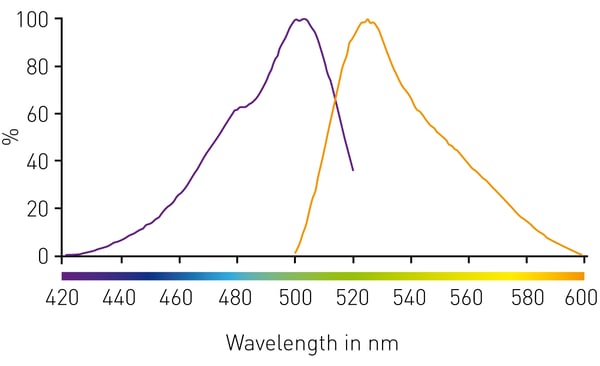
The Qubit® dsDNA HS is a fluorescent dye specific for double-stranded DNA and prevents measurement of contaminants such as RNA or aromatic impurities of DNA-extraction. To assess the settings for measuring the Qubit™ DNA dye, spectral scans for excitation and emission wavelengths have been performed using the CLARIOstar microplate reader (Figure 1). The excitation maximum was found to be at 503 nm and the maximum of emitted light at 525 nm, which reasoned to apply 485/520 filter combination or LVF MonochromatorTM settings of 483-14/530-30 for Qubit detection.
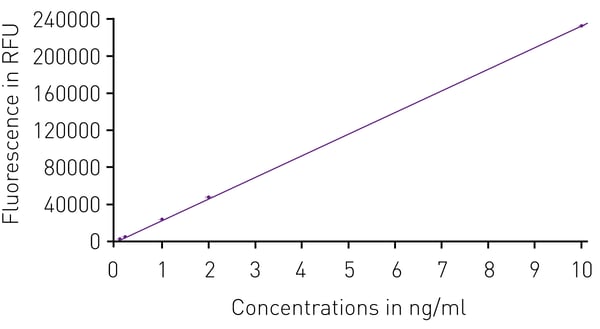
Next, the linearity of Qubit DNA quantification was tested. To this end, triplicates of Qubit standard DNA dilutions between 0.1 ng/µl-10 ng/µl were detected using the Qubit dye. The fluorescence signal was linear over these concentrations as indicated by a correlation coefficient R2 > 0.9999 (Fig.2). This linearity was obtained on FLUOstar Omega, CLARIOstar, and PHERAstar microplate readers.
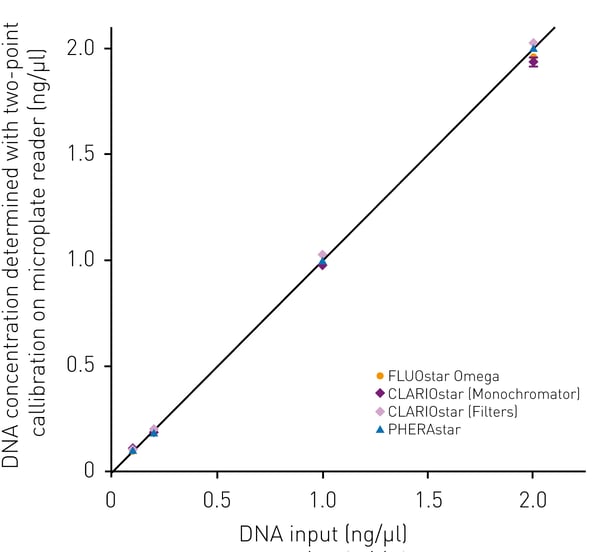
Given the linearity of Qubit DNA quantification in the concentration range between 0.1 ng/µl-10 ng/µl we hypothesized that in accordance with the Qubit measurement, two concentrations for defining a calibration curve might be sufficient. Quantifying known amounts of DNA with Qubit dsDNA HS chemistry and with a standard curve based on triplicates of the 0 and 10 ng/µl DNA standard, allowed to accurately determine the amount of DNA (Fig. 3). Taking into account that the sample volume may vary in the range between 1 µl and 20 µl as it is suggested by the manufacturer, the measurable DNA sample concentration range that can be quantified with Qubit on BMG LABTECH readers lies between 50 pg/µl-100 ng/µl.
Table 1: The table shows calculated DNA concentrations from two-point fit
| DNA concentration determined on BMG LABTECH microplate readers | |||
| Input DNA concentration | FLUOstar Omega | CLARIOstar | PHERAstar |
|
0.1 ng/µl
|
0.109
|
0.108
|
0.101
|
|
0.2 ng/µl
|
0.196
|
0.203
|
0.195
|
|
1 ng/µl
|
0.983
|
1.028
|
1.001
|
|
2 ng/µl
|
1.962
|
2.025
|
2.000
|
Conclusion
The use of the fluorescent Qubit dsDNA HS stain together with any multi-mode microplate reader by BMG LABTECH allows for the quantification of dsDNA samples with a concentration between 50 pg/µl-100 ng/µl. The combination increases throughput and enables easy replicate measurements.