Introduction
Influenza virus is a major threat to global public health. Essential to virus propagation is the viral RNA-dependent RNA polymerase (FluPol) which amplifies the viral genome and transcribes mRNAs coding for viral proteins. FluPol, a ~260 kDa multifunctional heterotrimeric complex, is an attractive target for anti-influenza drug discovery (1). Here, we present a novel fluorescence polarization assay that directly reads out RNA synthesis by FluPol (2). Moreover, the assay is applicable for measuring RNA synthesis in general and is compatible with high-throughput screening.
Assay Principle
The fluorescence polarization (FP) of fluorescently-labelled nucleic acids is sensitive to the conformation of the fluorophore’s environment. Specifically, the FP of FAM-labelled single-stranded RNA increases when it becomes double-stranded (Figure 1). Using an 18 nucleotide 5’-end FAM-labelled single-stranded “template” RNA, the highest signal amplitude (∆FPobs) is observed when adding 18 nt complementary RNA (black), resulting in a perfect 18 bp dsRNA. Increasing the single-stranded environment of the fluorophore by shortening the complementary RNA to 16 nt (yellow) or 14 nt (purple), respectively, reduced ∆FPobs max.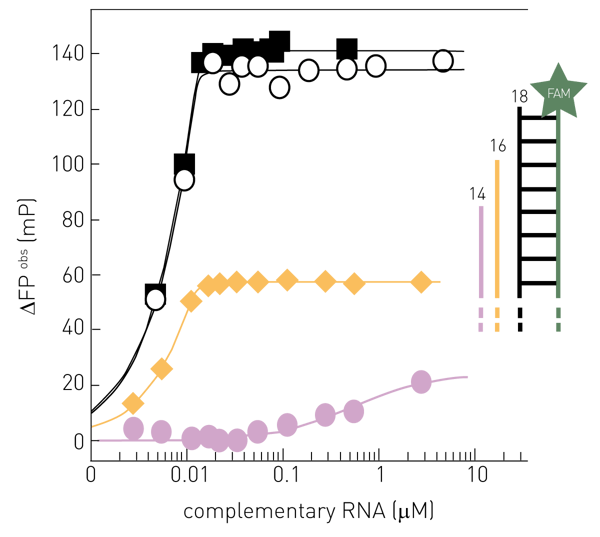
Active-site titration of the perfectly complementary RNAs of 18 nt (black) and 16 nt (yellow) was observed, while the interaction of the non-perfectly complementary 14 nt RNA (purple) would correspond to a KD of ~0.5 μM. Note that changing the RNA sequence or the hybridization strength may affect ∆FPobs max and shift the detection limit where the FP-signal is directly proportional to the amount of product RNA. The 5’-end FAM-labelled 18 nt “template” RNA used here actually corresponds to the influenza virus promoter 3’ RNA and this serves as template for RNA synthesis by FluPol in biochemical assays in vitro (2). This will produce a complementary 18 nt product RNA at a certain rate, which if it hybridizes to the FAM-labelled template will produce a change in its FP-signal. Before recording the FP-signal of the enzyme-catalyzed RNA synthesis, the reaction needs to be quenched (e.g. by high salt) to dissociate FluPol and RNAs while allowing RNA-RNA-interactions (Figure 2). With this “trick”, the contributions to the observed FP-signal of FluPol and the FAM-labelled template RNA interacting are removed and the detected FP-signal is directly proportional to the ratio of product RNA over template RNA (Figure 1).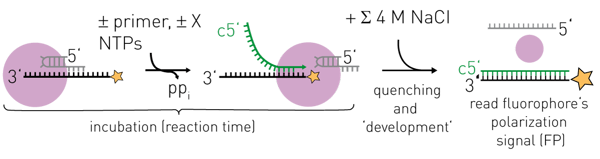
Materials & Methods
- Oligonucleotides (IBA Lifesciences)
- Primer (5’-(N7MeGppp)AAUCUAUAAUAG-3’), 2’F-2’dNTPs (Trilink Biotechnologies)
- NTPs (Sigma)
- FluPol (influenza B polymerase; Reich et al., 2014)
- 384 well plates (Greiner, #781076)
- CLARIOstar® (BMG LABTECH)
RNA synthesis was performed as described. Briefly, RNA synthesis was initiated by supplementing 0.25 μM FluPol (influenza B polymerase; bound to v5’ RNA nts 1-14) with 0.15 μM FAM-labelled template RNA (5’-(FAM-Ex-5) UAUACCUCUGCUUCUGCU-3’), 0.5 μM primer and 25 μM NTPs (each) in assay buffer (50 mM HEPES/NaOH, 150 mM NaCl, 10% (v/v) glycerol, 5 mM MgCl2, 2 mM TCEP, 1% (v/v) DMSO, pH = 7.4). RNA synthesis proceeded in polypropylene reaction tubes and at indicated times aliquots of 10 μl were transferred to 80 μl of quenching solution (4.5 M NaCl) before the FP-signal was recorded in 384 microplates using the CLARIOstar® (BMG LABTECH). Dr. J. Timmins (IBS Grenoble) is acknowledged for access to the microplate reader.
Instrument Settings
| Optic settings |
Fluorescence Polarization, endpoint |
|
|
Filters |
Ex: 482-16 |
|
|
Focus and gain |
adjusted before measurement |
|
|
Target mP |
35 |
|
|
Flashes |
50 per well |
|
|
General settings |
Settling time |
0.1 s |
Results & Discussion
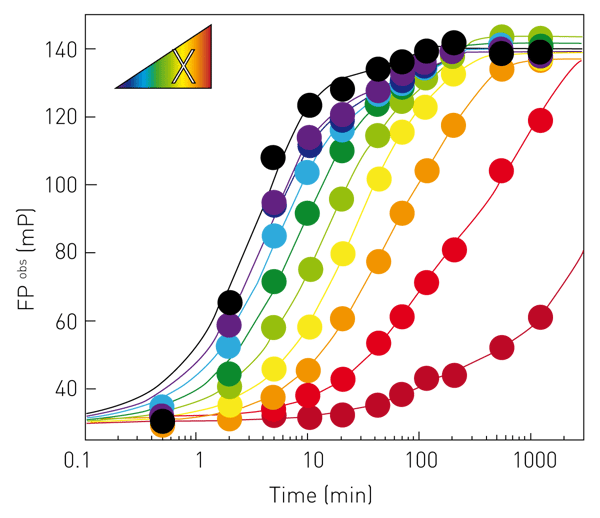
With the described RNA synthesis assay, influenza polymerase can be enzymatically characterized in vitro, e.g. regarding promoter RNA requirements, primer efficiency, KM (NTPs), turnover numbers, initiation strategies or inhibition (2). Kinetics of RNA synthesis are most informative and report on the amount of product and the rate it is produced. Figure 3 shows complete kinetics of RNA synthesis catalyzed by FluPol (primed with capped RNA) at 25 μM NTPs (black) and the effect of increasing concentrations of the inhibitor 2’F-2’dCTP (X; purple (8 μM) to dark red (8.3 mM)). Progress curves are fitted double-exponentially according to a pseudo-first order rate law (solid lines) enabling the RNA synthesis rate constants k (min-1) to be derived.
The rate constants corresponding to the prominent, fast RNA synthesis phase are shown in Figure 4, open black circles and yielded an IC50 (2’F-2’dCTP) of 0.2 mM. When quenching the synthesis reaction at a defined reaction time (within the linear range), the assay becomes compatible with high throughput screening campaigns. Figure 4 shows dose-response curves of NTP-analogues at conditions as in Figure 3 but allowing RNA synthesis to proceed for 5 minutes only and yielded e.g. IC50 (2’F-2’dCTP) = 0.2 mM (Figure 4, black squares), in agreement with the IC50-value determined from complete kinetics (Figure 4, black open circles).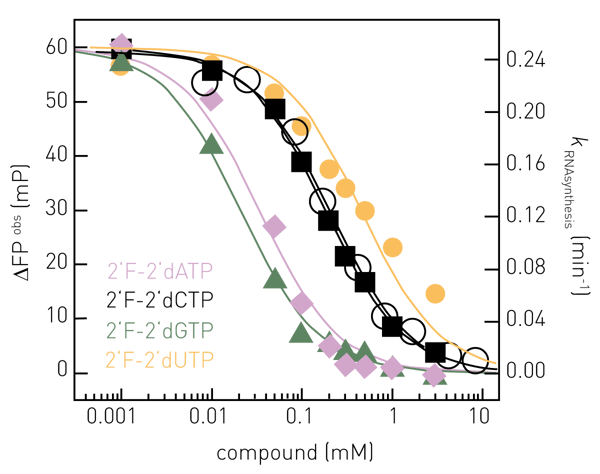
Conclusion
We developed a simple in vitro RNA synthesis assay that utilizes fluorescence-polarization changes of FAM-labelled model template RNAs associated with FluPol catalyzed product RNA formation. The assay is high throughput compatible and can easily be performed in 384 well microplates. It reliably reports on compounds inhibiting RNA synthesis, readily can be miniaturized to 0.2 pmole/reaction of recombinant (active) FluPol, and might provide an attractive choice for drug discovery campaigns.
References
- Pflug et al., Virus Res. 2017 Apr 15;234:103-117. doi: 10.1016/j.virusres.2017.01.013
- Reich et al., Nucleic Acids Research 2017 Apr 7; 45(6):3353-3368. doi: 10.1093/nar/gkx043