Introduction
Esterases catalyze hydrolysis reactions by converting esters into an acid and an alcohol using water as nucleophil. Therefore, esterases belong to the enzyme group of hydrolases. They are often used as biocatalysts to produce optically pure compounds.
Esterases differ in their affinity to specific substrates and this affinity is represented by the Michaelis-Menten constant Km. It describes the affinity of an enzyme to a substrate. Km is equivalent to the substrate concentration at which the reaction velocity is half of the maximal velocity. That means that a high affinity to a substrate leads to a small Km-value and vice-versa. BMG LABTECH has developed a new evaluation software feature able to calculate the Km value as well as the maximal velocity (Vmax) from an enzymatic kinetic measurement. Next to the Michaelis-Menten fit, other linearized fits are also shown and a comparison of the results is given.
As a model reaction, the p-nitrophenyl acetate (pNPA) assay was performed on a BMG LABTECH microplate reader.
Assay principle
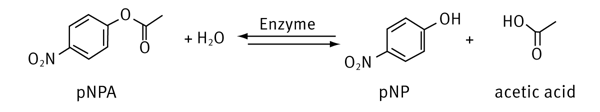 The enzyme hydrolyzes the acetate ester with the help of water. The products are acetic acid and p-nitrophenol (pNP), the latter showing an absorption maximum at about 405 nm.
The enzyme hydrolyzes the acetate ester with the help of water. The products are acetic acid and p-nitrophenol (pNP), the latter showing an absorption maximum at about 405 nm.
Materials & Methods
- p-nitrophenol (pNP) and p-nitrophenyl (pNPA) acetate, Sigma Aldrich
- Clear 96-well microplates, Greiner
- BMG LABTECH microplate reader
- MARS Data Analysis Software, Version 1.20 or higher
- Two different enzyme preparations (E1 and E2) were kindly provided by the working group of Prof. Bornscheuer, Greifswald
Enzyme kinetic measurements
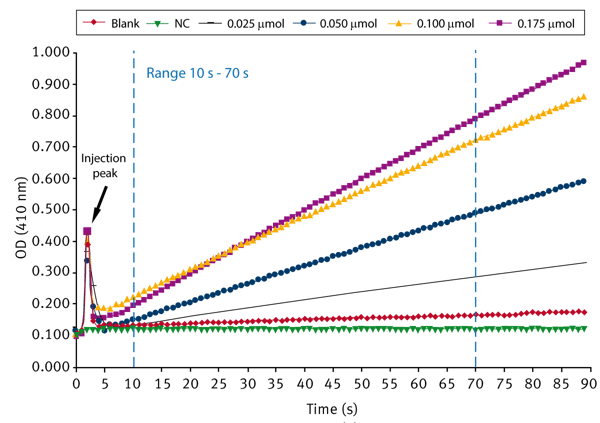
First a 10 mM stock solution of the substrate pNPA in DMSO is prepared. In each well 190 μL phosphate buffer (50 mM, pH 7.4) and 10 μL enzyme preparation is pipetted. 40 μL of substrate (solved in DMSO) at different concentrations is added using the onboard injectors. The final volume per well is 240 μL. The enzymatic reaction starts after adding the substrate. The absorbance is measured at 410 nm for every second for 90 seconds at 37°C. The blank represents the autohydrolysis of pNPA without enzyme. In addition, a pNPA-free negative control (NC) is run that consists only of buffer and enzyme.
The measurement is performed using the well mode (see instrument settings below) that provides the possibility to inject and to measure absorbance at the same time.
Instrument settings
| Measurement type: | Absorbance, Well Mode |
| No. of intervals: | 90 |
| Kinetic interval time: | 1 s |
| No. of flashes per well: | 20 |
| Positioning delay: | 0.5 s |
| Target temperature: | 37°C |
| Filter: | 410-10 nm |
Injection settings
| Volume of pump 1 and 2: | individual |
| Pump speed: | 310 μl/s |
| Injection start time: | 2.0 s |
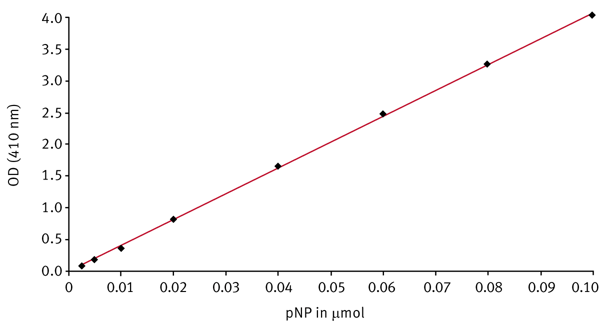
 pNP standard curve
pNP standard curve
For the calculation of the initial reaction rates it is necessary to convert the delta OD values per minute into μmol product per minute. Therefore a standard curve of the product using the same reaction conditions as for the kinetic measurements has to be done. From a 10 mM pNP stock solution, solved in phosphate buffer (50 mM, pH 7.4), different dilutions of pNP ranging from 0.0025 μmol to 0.1 μmol were prepared in 200 μL. 40 μL of DMSO were added simulating the DMSO concentration in the kinetic samples. The optical density (OD) is measured using an absorbance 410-10 nm filter. Figure 3 shows a pNP standard curve.
The slope of the standard curve stands for the extinction coefficient for pNP under the conditions mentioned. It was determined to be about 41 OD per μmol. The coefficient is used to calculate the initial reaction velocities.
Results & Discussion
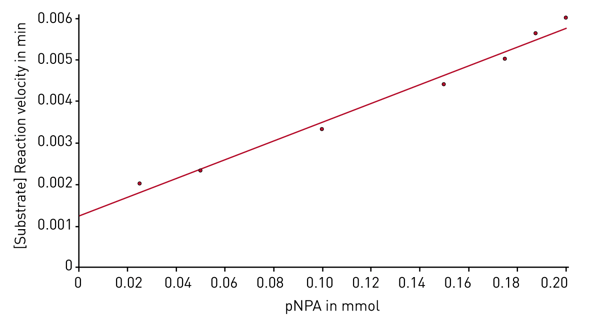
After the enzyme kinetic measurement is finished, the evaluation software provides different plots and presents the corresponding Km and Vmax values. The Michaelis-Menten fit shows the reaction velocities depending on the substrate concentration (Figure 4).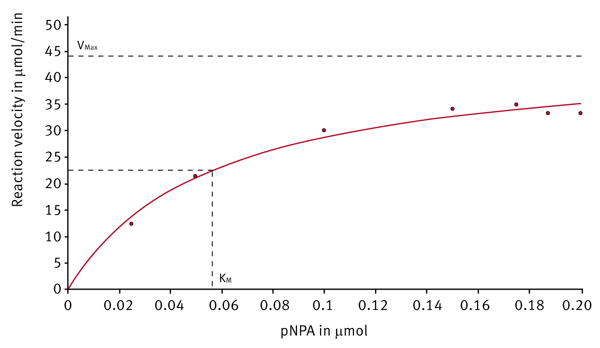 Next to the Michaelis-Menten fit different linearized plots are available. Figure 5 shows such a linearized diagram (Hanes plot).
Next to the Michaelis-Menten fit different linearized plots are available. Figure 5 shows such a linearized diagram (Hanes plot).
The results of further linearized fits are compared to the Michaelis-Menten findings in table 1.
Table 1: Km and Vmax comparison obtained from different enzyme kinetic plots.
| Plot | E1 |
E2 |
||
| Km [μmol] | Vmax [μmol/min] | Km [μmol] | Vmax [μmol/min] | |
| Michaelis-Menten | 0.056 | 44.8 | 0.12 | 16.3 |
| Lineweaver-Burk | 0.073 | 49.2 | 0.08 | 12.5 |
| Eadie-Hofstee | 0.063 | 46.4 | 0.07 | 13.2 |
| Scatchard | 0.068 | 47.9 | 0.10 | 15.2 |
| Hanes | 0.056 | 44.4 | 0.11 | 15.5 |
The different methods to determine Km and Vmax have all assets and drawbacks. Because of that, it is recommended to have a look at different plots for deciding which calculated constants are really correct. With the help of the MARS Data Analysis software enzyme, kinetic evaluation is very simple because every plot is available directly after measurement and calculation.
Conclusion
The new software feature for enzyme kinetic offers fast and easy calculation of Km and Vmax. Available plots are the common Michaelis-Menten, Lineweaver-Burk, Eadie-Hofstee, Scatchard and Hanes kinetic fits.